Research Projects
Our research investigates how organisms adapt to their environment, with a focus on the evolution of complex traits. We test hypotheses on ecologically-mediated selection in species with relevance to human health: diverse human populations, non-human primates, and vectors of important diseases.
Disease immunogenetics in diverse human groups:

Major priorities of our research group include infectious diseases that cause the most excess and inequitable childhood mortality. Two such diseases are malaria and lower respiratory tract infections. In these projects, our objective is to understand the genetic and immunological reasons why human populations with different ancestral subsistence strategies or different mosaics of genomic ancestry differ in their present disease burden and host-pathogen interaction dynamics. We use high-throughput transcriptomic, genomic, serological, and microbiome methods to understand the diseases in an evolutionary and anthropological framework. We work in parts of tropical Africa (Uganda and Madagascar) with the highest morbidity and mortality for these diseases, and we prioritize partnership with participant communities, as well as collaboration and capacity building with in-country scientists and clinicians. I have been fortunate to previously pursue evolutionary research at these study sites (e.g., Bergey et al. 2018, PNAS; Harrison et al. 2019, Nature Ecology and Evolution).
Collaborators include: Bwindi Community Hospital, The Batwa Development Program, Action for Batwa Empowerment Group, Sylvia Kokunda, Centre ValBio, Jason Hodgson, Rindra Rakotoarivony, and Andrea Manica.
Malaria mosquito evolution:

Human disease risk is also impacted by vectors of infectious diseases. With this motivation, we also investigate natural selection in mosquitoes that transmit malaria (genus Anopheles). My past work has focused on connectivity between populations, most prominently between islands in Uganda put forth as potential pilot sites for vector control with genetically modified mosquitoes (Bergey et al. 2019, Evolutionary Applications; Lukindu et al. 2018, Malaria Journal; Wiltshire et al. 2018, Parasites and Vectors; Miles et al. 2017, Nature). International work delayed by the pandemic focuses on ecological adaptation of mosquitoes to marginal habitats predicted to undergo rapid climate change and the genetic basis in shifts to zoophagic and exophagic behaviors, i.e. those that feed on non-humans, outdoors.
Current research also focuses on the mosquito immune response to pathogens (Henderson et al. 2022, PLOS Neglected Tropical Diseases; Henderson et al. 2022, G3) and our work manipulating RNA-interference (RNAi) in the mosquito immune system (summarized below).
Collaborators include: Jonathan Kayondo, Martin Lukindu, Bernadette (Menja) Rabaovola, Centre ValBio, and Rachel Wiltshire.
Gene drives based on eliciting piRNA biogenesis in insect pests:
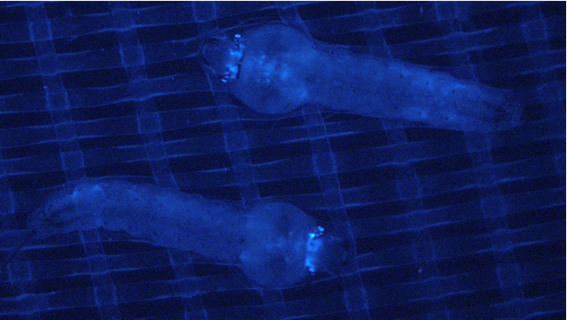
In the laboratory, we have created genetically modified insects to better understand the powerful piRNA pathway of the insect immune system. We have developed a method that capitalizes on this pathway to drive a desired trait to high frequency in a population of insects, such as those that vector disease or feed on crops. As proof of concept for our novel approach, we have generated transgenic mosquitoes with reduced ability to transmit malaria. Our method has been awarded a provisional U.S. patent and is available for license via Rutgers Research.
Besides blocking transmission of human pathogens in vector species like mosquitoes, we are particularly excited about the prospects of this method for control of crop pests, which create a large economic and social impact through direct damage to plants and spread of pathogens. For instance, some estimates suggest a single pest, corn rootworm, causes $1 billion in damage annually in the U.S. alone. Methods to combat insect pests have primarily relied on pesticide applications, which can have negative environmental and health impacts.
Collaborators include: Cory Henderson, Marcelo Jacobs-Lorena, and Rob Harrell.
Non-human primate evolution:

For my dissertation (Bergey et al. 2016; PNAS), I studied a natural experiment in hybridization, in which two species of wild baboons produce fertile offspring in Ethiopia. I found that regions of high differentiation between the species are more likely than expected to contain genes that are components of the dopamine receptor mediated signaling pathway, suggesting adaptive divergence between the two primates in brain regions involved in impulse control and linked to mating behavior.
Current primate projects are myriad and often driven by graduate and undergraduate students. These include an investigation of primate response to malaria (Ph.D. student Amber Trujillo; Trujillo and Bergey preprint), of sexual selection on sperm-associated genes (Ph.D. student Becca DeCamp), and of lemur response to anthropogenic impacts (Ph.D. student Lindsey Hauff).

Major ongoing primate projects focus on the dynamics of gene flow and selection from a phylogenomic perspective. Such NSF-funded projects include: 1.) an investigation of hybridization between members of a specious group of diverse African primates, the guenons, and 2.) an investigation of evolutionary conservation in anthropoid (monkey and ape) genomes and its application to distinguish false positives from causal variants in human disease association studies.
Overall, our primate research includes work on adaptation (Chiou et al. 2022, Nature Ecology and Evolution), hybridization (Rogers et al 2019, Science Advances; Chiou et al. 2021, Molecular Ecology) and genomics from non-invasive sources, including museum specimens and feces (Burrell, Disotell, and Bergey 2015; Journal of Human Evolution; Chiou and Bergey 2018; Scientific Reports; Marciniak et al. 2021; PNAS).
Collaborators include: Amber Trujillo, Becca DeCamp, Lindsey Hauff, Anthony Tosi, Luca Pozzi, Andrew Burrell, Clifford Jolly, Todd Disotell, Kenny Chiou, Jane Phillips-Conroy, Jeff Rogers.
